Biodiversity Monitoring
Using Field-Friendly Genetic Tools to Monitor Species
Tropical biodiversity faces unprecedented threats today, with species extinctions occurring faster than species documentation and characterization in some regions. Genetic tools for biodiversity monitoring go hand-in-hand with traditional monitoring tools (foot surveys and automatic field cameras) and more modern techniques (acoustic and drone-based surveys). But many questions as yet remain unanswered in the application of these tools to tropical ecosystems.
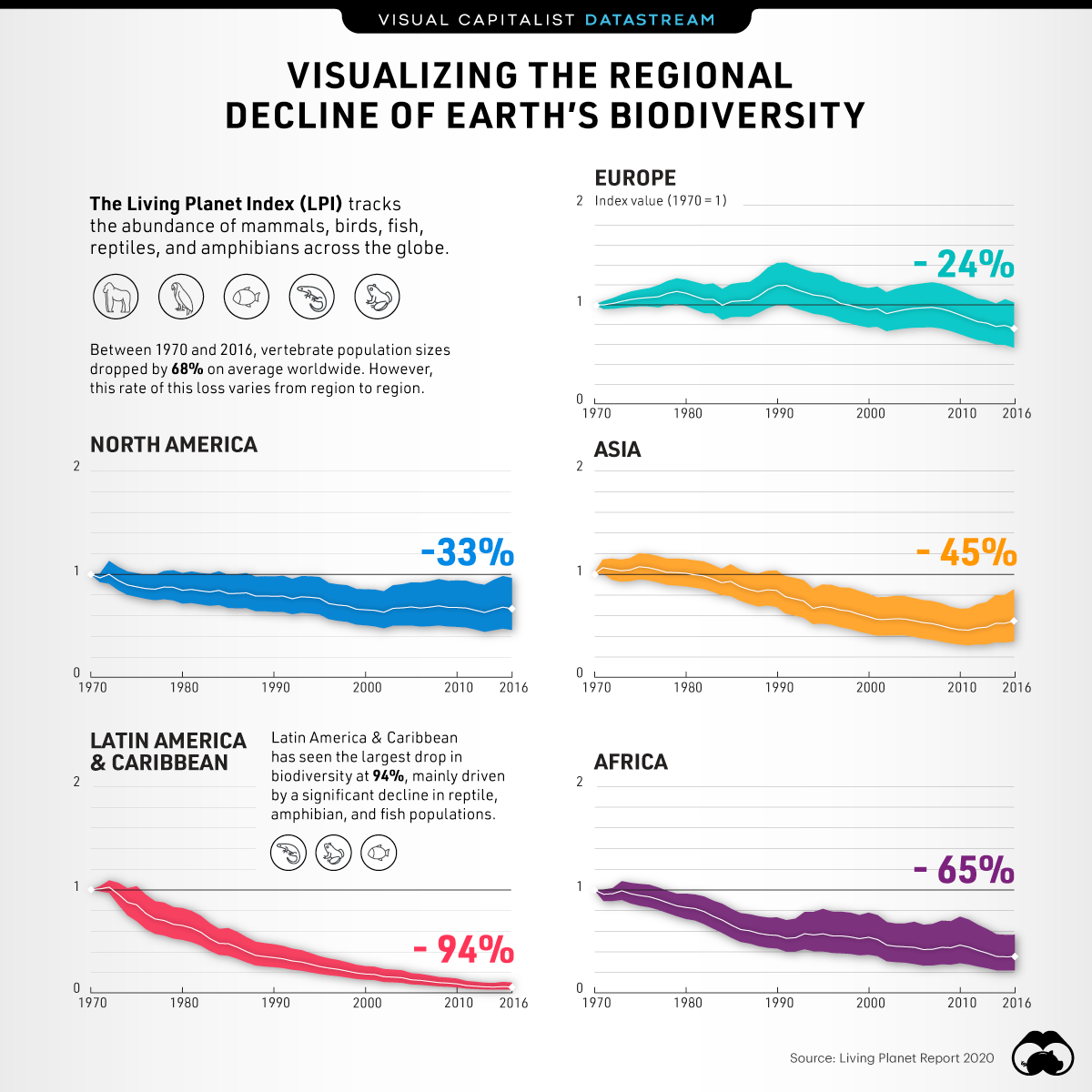
The world’s biodiversity is under threat – that much is undeniable. The primary threats to this biodiversity are related to humans: how we use land, extract resources, and dispose of waste have created myriad downstream threats to global biodiversity. As a result of this, the last 50 years have seen massive declines in populations, based on the Living Planet Index Report as visualized below. Stopping or slowing the rates of decline are of primary importance, but so is documenting the effect of such interventions to be able to choose actions with the most impact on biodiversity conservation. However, to do so, one must employ a toolset focused on describing biodiversity in a standardised, comparable, and reliably accurate manner.
How Labs Save Biodiversity
The ISL Initiative aims to create a blueprint for sustainable, decentralized, and low-cost laboratories, that —rather than replacing existing urban facilities— will expand laboratory analysis capacity into areas that have historically lacked access to such resources. The capacities vital for this project include the ability to conduct DNA barcoding, high-throughput sequencing, DNA metabarcoding, genotyping, and environmental DNA (eDNA) monitoring.
DNA Barcoding
Over the last two decades, genetic sequencing of universal DNA barcodes has allowed researchers without taxonomic training to identify species from nonlethal, noninvasive and environmental samples using large, open-access DNA reference repositories. However, the production of species DNA barcodes still lags in many countries that boast the highest biodiversity, in large part because in-country access to sequencing services, infrastructure, and funding are lacking.

The Barcode of Life Database, or BOLD, was launched in 2007 as an “an informatics workbench aiding the acquisition, storage, analysis and publication of DNA barcode records.” Animals (marker name COI), plants (RBCL and MATK), and fungi (ITS) sequences are curated across multiple taxonomic groups alongside precise morphological and geographic data. There is no better place to deposit sequences at this time, and our work will be hosted as an open access resource (link to be announced shortly).
DNA Metabarcoding
DNA barcoding has traditionally relied on Sanger sequencing, where each genetic marker for a particular sample is sequenced independently. Sanger sequencing, however, relies on equipment that is large, expensive, and not easily accessible. Barcoding efforts have responded primarily in one way to this challenge: by providing pipelines for the export of samples to large sequencing hubs. However, this continues to remove analysis pipelines from the hands of the scientists in many countries that are in critical need of local sequencing solutions, and place them in the hands of overdeveloped nations instead.
Metabarcoding is the simultaneous sequencing of multiple copies of a gene, for example from an environmental sample. Metabarcoding requires the use of high throughput sequencers. Examples of metabarcoding include diet analyses of fecal samples (18S or 12S markers, e.g.), microbial metabarcoding in fecal or soil samples (16S markers, e.g.), or environmental DNA analyses of water samples (12S or 18S markers, e.g.).
High-throughput Sequencing
Traditionally, DNA barcoding has been conducted on one gene per sequencing reaction, using Sanger sequencing (or a first generation sequencing method). However, as genomic sequencing has increased in efficiency and scale, second and third generation sequencing technologies have emerged. They range from high throughput sequencing allowing for entire genomes to be sequenced in a day to single molecule sequencing using nanopore technology. Note, the term “next-generation sequencing” is now synonymous with second-generation sequencers. Now that a third generation has arisen, we are more open to the idea that there might be more generations to come.
Metagenomics
Metagenomics is the study of sequences recovered directly from an environmental sample, without targeting of any specific genes. This is often applied to microbial communities, but it can also be used to discover viruses, assemble genomes, and much more. Practically speaking, metagenomics is when you take a sample, extract whole genome DNA from it and sequence all of that DNA directly. Some methods can be applied right before to enrich for bacterial, viral or host genomic DNA, if you have a specific goal in mind. Metagenomics is useful in assembling metagenomes, or full genomes of organisms contained in a mixed environmental sample.
Metagenomics therefore represents the metabolic potential of your sample. To know if these genes are indeed being expressed, then you would need to analyse the transcriptome, or mRNA in your sample at the time of sampling.
eDNA
DNA can exist in the environment for a surprisingly long period of time. In a given soil sample, there is DNA from animals, plants, fungus and bacteria in the environment, at a minimum. Environmental DNA is broadly described as congruent with noninvasively collected samples, and therefore includes DNA from fecal, soil, water, or even air samples.
eDNA typically utilizes a metabarcoding approach by assessing a few targeted genetic markers and sequencing them simultaneously out of environmental samples on a high-throughput sequencer (all of the terms underlined were defined on this page). It is therefore a powerful tool to explore a new environment, compare microhabitats, compare the effects of resource extraction on an environment, and more. However, since animals deposit DNA into the environment at different rates, and DNA degrades differentially based on environmental conditions, standardizing eDNA analyses for specific cases is a must. Although eDNA can quite confidentially verify if a species is presence or not, it takes a very standardized and validated protocol to actually infer how much DNA of one species is present in the sample, compared to another species.
Trace DNA
As the study of environmental DNA has developed, a subset of samples have emerged that share some additional specific features. Typically, environmental DNA produces a DNA soup containing many different species’ DNA in a single sample. This remains at species-level identifications, because it is possible that more than one individual of a single species is also represented in the same samples. When more than one animal of a species has introduced DNA into a sample, it is nearly impossible to tease each genome apart. However, a small subset of samples does give us an opportunity to pull individual level DNA out of a sample, and we call this trace DNA.
Trace DNA is therefore DNA derived from samples deposited in the environment that can be linked back to a specific individual of a species. These include footprints, urine sprays and fecal material. Trace DNA from footprints has been primarily explored in frozen environments, where a sample of snow can be melted, and DNA retrieved from the water sample. However, we can indeed explore trace DNA from footprints in other environments as well.
Header Photo Credits:
Claudia Rohr (Otter), Ryan Peters (Bats), Timothy Paine (Frog)

